VoCamp1/Proposal
This is the original proposal turned into NESCent, which subsequently decided to sponsor the lion's share of the participant support costs.
Proposal for a Phyloinformatics VoCamp
Co-leaders. The group of organizers preliminarily consists of N. Cellinese, K. Cranston, H. Lapp, E. Pontelli, Sheldon McKay, A. Stoltzfus, and R. Vos, and is chaired by A. Stoltzfus (corresponding PI).
Synopsis. Controlled vocabularies and ontologies are key to the ability to integrate and compute over data across fields, experimental systems, and protocols. Ensuring that such standardized interoperability artifacts continuously evolve to meet changing research demands requires an open, community-based development process. At a recent NESCent-sponsored hackathon on Evolutionary Database Interoperability, ontologies and vocabularies that meet the needs of diverse community resources and tools emerged as a key gap. Filling this gap on a sustainable basis requires a diverse community of domain experts, users, and stakeholders with a shared awareness of, and commitment to, knowledge-based standards. To begin building such a community, we propose a "VoCamp"-style meeting for investigators to create and develop ontologies and lightweight vocabularies in support of integration and semantic cross-linking of evolutionary data with its many related fields. To take advantage of such cross-initiative synergy, and to foster better communication and knowledge exchange between them, we propose to co-localize the event with the annual meeting of the International Biodiversity Information Standards Organization (TDWG).
Background. The work of the Evolutionary Informatics Working Group at NESCent on addressing obstacles to interoperability of evolutionary data and tools has given rise to a "stack" of emerging standards for transmitting and accessing data and its semantics in a reliable and programmable way. Specifically, the stack consists of a rigorously defined and validatable syntax standard for phylogenetics data and trees with embedded and semantically rich metadata (NeXML), a standard programmable interface to online phylogenetic data resources (PhyloWS), and an ontology defining the terms and concepts used in comparative evolutionary analysis and the relationships between those (CDAO).
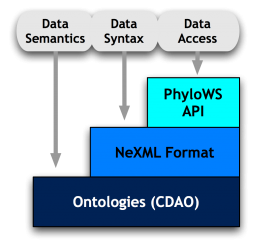
The working group concluded its work in March 2009 with a hackathon event focused on interoperability of evolutionary data resources. The event was successful in multiple ways. First, it produced working prototypes demonstrating interoperability improvements based on the EvoIO stack. Second, it brought together stack developers with experts from key online data resources, increasing awareness of interoperability obstacles as well as preferred approaches to overcoming those obstacles. The hackathon showed that, with a modest amount of training and effort, data providers can use the stack to improve interoperability, with benefits for data providers and for end users. Many participants have remained in contact in order to extend the work begun at the hackathon. A group of participants collaborated to propose an EvoIO data interoperability network in response to NSF's call for community-based INTEROP proposals. If successful, this would provide major funding for a community-based interoperability initiative.
The hackathon also exposed limitations. Participants repeatedly identified a gap in the availability of a sufficiently expressive ontology, or set of ontologies and vocabularies, along with community mechanisms and infrastructure to sustain their continued development and evolution. This was confirmed by several subsequent efforts to adopt some of the standards, for example incorporating PhyloWS and NeXML-support into the next-generation version of the community resource TreeBASE.
Motivation: Need to extend ontologies. Online repositories of evolutionary data contain data and metadata that far exceed the scope of the current CDAO ontology. The overall goal of the EvoIO stack is to allow this data to be accessed, searched, retrieved, and repurposed programmatically. The NeXML and PhyloWS projects allow describing, sharing, and querying data from diverse online databases, but their development is being hampered by a lack of controlled vocabulary and ontologies necessary to transport the semantics in an interoperable manner. Examples that have already been identified through the work of hackathon and EvoInfo participants include metadata on experimental protocols, ontology-based annotation of phenotypes, and taxonomic affiliations of OTUs.
Even if the EvoIO stack projects agree on terminology, developing sustainable solutions to these problems requires coordination and direct involvement of a much larger community of stakeholders. A sustainable solution requires nurturing a community dynamic in which domain experts, users, and developers alike know the value of shared terminology, and know how to contribute to its development.
We propose to jump-start such a community dynamic by holding a VoCamp-style event. A VoCamp is similar in conception to a hackathon in the sence of being an intense, hands-on, working meeting with face-to-face interactions between a diverse group of people who create an intellectually fertile stimulating atmosphere. Instead of developing software source code, it focuses on issues of vocabulary and ontology design, development, and application. VoCamps have emerged only recently (in 2008), but have since spread rapidly (see the online list of upcoming and previous VoCamps), and hold significantly potential to foster collaborative and open development and community cohesion for ontologies as hackathons have been shown capable of for open-source software development.
Meeting preparation. To maximize the productivity of the event, it is important that those who attend are committed to, and capable of, making substantive contributions to the development process, and that the participants represent a balanced mix of ontological engineers, logicians, domain experts, as well as developers who would consume and apply the ontologies.
We propose to hold this meeting co-incident with the 2009 TDWG Annual Meeting in Montpellier, France. This would allow us to take advantage of opportunities for synergy with the recent shift of TDWG activities towards shared vocabularies and ontology development, as well as for broadening the nascent EvoIO community to biodiversity informatics practitioners, which includes information scientists working in museums, ecology, and conservation. A European location would also be very cost-effective for some of the CDAO developers, who are based in Strasbourg, and for involving the Bio-Health Informatics and the Information Management Groups at the University of Manchester, which comprise some of the world's leading ontologists and semantic web experts. We expect the TDWG-related venue would be attractive to a variety of initiatives including Darwin core, Biological Descriptions, Taxonomic names and concepts, GBIF, EOL, Observational Data, and DataOne. Aside from these opportunities to connect disparate communities facing similar issues, holding the event at the TDWG meeting would double as an ongoing activity of the TDWG Phylogenetics Standards Interest Group, which is co-led by two members of the organizing group (Lapp and Cellinese).
Additional participants will be recruited through an open call for participation disseminated to data providers, mailing lists for the EvoIO Stack, the Scientific Observations Network (SONet), TDWG mailing lists, GBIF, the Society for Systematic Biology, and the EvolDir and EcoLog mailing lists.
Meeting agenda. As successfully practiced previously for the NESCent-sponsored hackathons, the exact agenda will be developed by the participants, partly in advance through teleconferences and online electronic media, and partly on-site through an Open Space activity. We anticipate, however, that the following activities and tasks will be needed to ensure a productive event for all participants.
- Boot-camps on ontology design and engineering, ontology languages and their restrictions, entailment and reasoning over ontologies, and infrastructure for collaborative development of ontologies.
- Identification of focus areas for vocabulary development and term definition.
- Identification of targets of opportunity for cross-project and cross-community synergy.
Similar as in hackathons, the participants will break into smaller subgroups that collaboratively work on their chosen development targets. The total event duration is envisioned to be between 4 and 4.5 days. The event will conclude with a wrap-up session, and if held at the TDWG Annual Meeting, a report will be scheduled to be presented afterwards to the TDWG Conference audience.
Expected outcomes. We expect that the event will have a substantive impact on multiple ontology development efforts, stakeholder communities, and interoperability initiatives, by connecting previously disparate communities with shared objectives, sharing knowledge, and by actually building out existing ontology resources in terms of ontological rigor, semantic richness, and modularity that supports effective reuse. Specifically for evolutionary data interoperability, we expect the event to result not only in a much richer and well-defined CDAO that meets the immediate needs of online data providers and aggregators, but also in a much improved alignment of CDAO design principles with initiatives from ecology (such as SONet), biodiversity (such as DarwinCore and the TDWG Ontology), and genetics / biomedicine (such as OBO).
We anticipate that the event, if successful, will give rise to similarly structured follow-up events organized by these other communities; although subject-focused ontology development sprints have taken place for OBO ontologies (such as for specific regions of an anatomy), more inclusive VoCamp-style events have not yet in any of those. If the EvoIO Community Network is funded, the expected outcomes of this event will provide the network with a substantial head-start both in terms of standards development as well as community diversification and engagement.