Phylotastic/Architecture
Screencast
http://www.youtube.com/watch?v=d-fDngweW-M
Overview
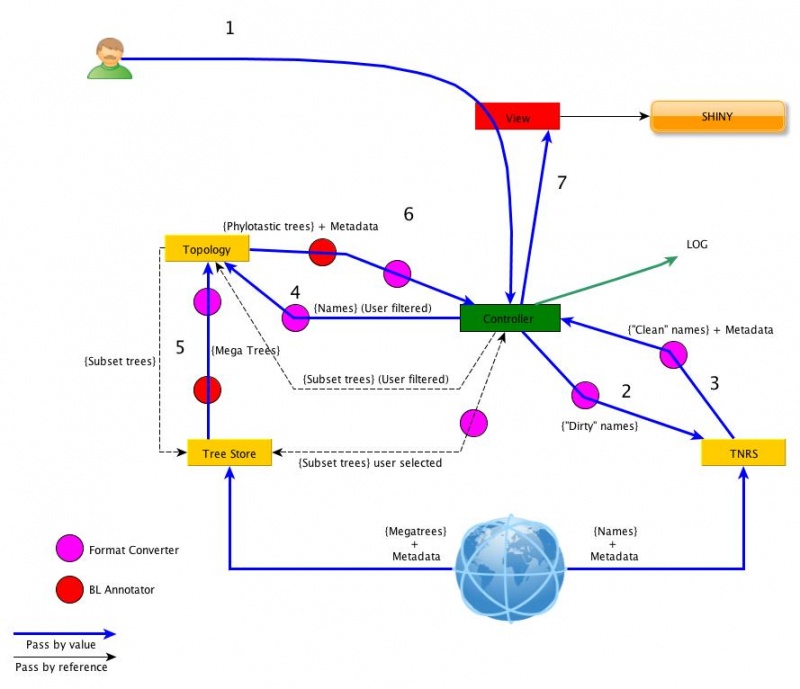
Our architecture is based on the Model-View-Controller design pattern and takes the following modules into consideration:
- TNRS
- Topology prune/graft
- Tree Store
- Syntax Format Converter
- Branch length annotator
- Logger
The architecture diagram below presents the interaction between the various modules. Each module will be treated as a "black box" in this architecture and the only elements we are concerned about here will be the interoperation between the modules.
These should fuel the input/output specifications for each module. The architecture also clarifies what is passed by reference (blue arrows) from what is passed as values (black arrows). As an example, mega trees should not be passed directly to the controller but, instead, only the reference to those mega-trees should be sent. These references are then passed to the topology module, which will use those trees.
Steps:
- Start
- list of name strings [mandatory user input]
- TNRS sources
- TNRS “knobs” (“fuzziness” etc.)
- Post TNRS
- Choices about unresolved names
- Megatree retrieval
- (mega-)tree store source
- automated tree query for applicable trees (per treestore) with user-supplied parameters
- (mega-)tree filter/selection criteria (e.g. degree of overlap)
- Pre-Topology
- application of branch lengths?
- Topology
- (see Tolopolgy, below)
- Post-topology
- application of branch lengths?
Sample Workflow
Input
Input species list: "Homo sapiens" (human), "Pan troglodytes" (chimp), "Gorilla gorilla" (gorilla)
Step 1: TRNS Resolution
Invocation:
<TRNS_SERVICE_URL>?query=Homo+sapiens%0APan+troglodytes%0AGorilla+gorilla
Output (JSON):
{
"metadata": {
"jobId": 1,
"submitDate": "2012-06-06T14:54Z",
"sources": [{
"sourceId": "ITIS",
"sourceName": "Integrated Taxonomic Information System",
"uri": "http://www.itis.gov/",
"rank": 1,
"status": "online",
"annotations": {"TSN": "Taxonomic Serial Number, ITIS' internal identifier"}
}, {
"sourceId": "NCBI Taxonomy",
"sourceName": "NCBI Taxonomy",
"uri": "http://www.ncbi.nlm.nih.gov/taxonomy",
"rank": 2,
"status": "online",
"annotations": {'nucleotide_uri': "A link to nucleotide sequences on GenBank for this taxon", 'protein_uri': "A link to protein sequences on GenBank for this taxon."}
}]
},
"names": [{
"submittedName": "Homo sapiens",
"matchCount": 2,
"matches": [{
"sourceId": "ITIS",
"matchedName": "Homo sapiens",
"acceptedName": "Homo sapiens",
"uri": "http://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value=180092",
"annotations": { "TSN": "180092" },
"score": 1.0
},
{
"sourceId": "NCBI Taxonomy",
"matchedName": "Homo sapiens",
"acceptedName": "Homo sapiens",
"uri": "http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=9606",
"annotations": { },
"score": 1.0
}]
},
{
"submittedName": "Pan troglodytes",
"matchCount": 2,
"matches": [{
"sourceId": "ITIS",
"matchedName": "Pan troglodytes",
"acceptedName": "Pan troglodytes",
"uri": "http://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value=573082",
"annotations": { "TSN": "573082" },
"score": 1.0
},
{
"sourceId": "NCBI Taxonomy",
"matchedName": "Pan troglodytes",
"acceptedName": "Pan troglodytes",
"uri": "http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=9598",
"annotations": { },
"score": 1.0
}]
},
{
"submittedName": "Gorilla gorilla",
"matchCount": 2,
"matches": [{
"sourceId": "ITIS",
"matchedName": "Gorilla gorilla",
"acceptedName": "Gorilla gorilla",
"uri": "http://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value=573080",
"annotations": { "TSN": "573080" },
"score": 1.0
},
{
"sourceId": "NCBI Taxonomy",
"matchedName": "Gorilla gorilla",
"acceptedName": "Gorilla gorilla",
"uri": "http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=9593",
"annotations": { },
"score": 1.0
}]
}]
}
Step 2: Identify Candidate Trees
Invocation: HTTP POST the following JSON to <TREE_STORE_URL>/phylows/find/tree
taxa_uris=[
"http://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value=180092",
"http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=9606",
"http://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value=573082",
"http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=9598",
"http://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value=573080",
"http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=9593"
]
This JSON is extracted from the output of the TNRS service.
Output: URIs of matching trees in the tree store +Provenance
{
"http://example.com/tree10" : { "label" : "something", "author": "", "matches" : ["http://example.com/taxa1"] },
"http://example.com/tree34": { "label" : "something", "author": "", "matches" : ["http://example.com/taxa2"] }
}
Step 3: Pruning Service
Invocation: HTTP POST the following JSON to <PRUNING_SERVICE_URL>
tree_uri=http://www.evoio.org/wg/evoio/images/2/26/Bininda-emonds_2007_mammals.nex taxa_uris=[ "http://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value=180092", "http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=9606", "http://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value=573082", "http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=9598", "http://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value=573080", "http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=9593" ]
Output: Pruned tree
While processing the above request from the controller, the pruning service will retrieve the full representation of the tree from the tree store:
Invocation: HTTP POST the following to the <TREE_STORE_SERVICE_URL>/tree
tree_uri=http://www.evoio.org/wg/evoio/images/2/26/Bininda-emonds_2007_mammals.nex
Output: Full tree in NeXML or Newick. For example:
Interface description language example
<tool id="fa_gc_content_1" name="Compute GC content">
<description>
for each sequence in a file
</description>
<command interpreter="perl">toolExample.pl $input $output</command>
<inputs>
<param format="fasta" name="input" type="data" label="Source file"/>
</inputs>
<outputs>
</outputs>
<tests>
<test>
<param name="input" value="fa_gc_content_input.fa"/>
<output name="out_file1" file="fa_gc_content_output.txt"/>
</test>
</tests>
<help>
This tool computes GC content from a FASTA file.
</help>
</tool>
Topology Module
_per megatree in:_
Input
- List of names [mandatory; post-cleaning]
- with Taxonomic cues/guides [optional; can also be auto-discovered]
- Megatree choice
- Configuration:
- Grafting policy
- choices of insertion of non-matching terminals
- sister to random terminal
- random node in matching clade
- basal
- conservatively collapse clade
- choices of insertion of non-matching terminals
- Pruning policy
- restrict tips to names given, or return all tips in minimum spanning clade
- retain or delete out-degree one nodes
- how to handle metadata associated with nodes/edges that have been deleted
- Grafting policy
API
- TNRS: please see API in TNRS
- treestore
Request:
taxa_uris=http://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value=180195%0Ahttp://www.tropicos.org/Name/1300071
Results:
{
"http://example.com/tree10" : { "label" : "something", "author": "", "matches" : ["http://example.com/taxa1"] },
"http://example.com/tree34": { "label" : "something", "author": "", "matches" : ["http://example.com/taxa2"] }
}
Output
- One or more tree(s)
- topology
- node-by-node metadata [as per megatree, optional]
- list of non-matching taxa
- logging record to logger
Branch lengths
Possible strategies:
- NPRS
- Based on input BLs
- Default (no input BLs)
- Node-age constrained equidistant adjustment (BLADJ)
- refer to BL group (Congruifier)
Topology Scenario and Dependencies
Topology services depend on two other services: TNRS and TREE SOURCE User starts by submitting a list of names and a set of configuration elements (e.g. sources to use, knobs, etc). TNRS returns list of taxa and their URI. User then decides whether resolved names are correct. This list of chosen URI taxa will be sent to the tree source, along with a selection of mega tree sources. The tree store returns the URI of a set of megatrees that are applicable for the taxa submitted. The user selects, from these trees, the list of trees that he/she wants to use. This list, along with the taxa uri, is submitted to topology module, along with a set of configuration instructions. Phylotatic tree (or trees) are returned to the user, including node-by-node metadata information.
Controller Behavior
Error creating thumbnail: Unable to save thumbnail to destination